10 January 2022
Fast, contiguous and accurate crop genome assembly (arabidopsis (Col-0) and tomato (Heinz 1706), thanks to new chemistry, nanopores and plant-aware basecaller
We would like to announce the release of the first crop genome assembly datasets from Arabidopsis and Tomato, based on the latest Oxford nanopore R10.4 flow cells combined with the Q20+ chemistry generated at KeyGene. The release encompasses the pass-filter simplex data (.fast5 format, QV10) and the reference genome assemblies (.fasta) that have been generated. Highest raw read accuracies were obtained by using a plant-aware basecalling model, which resulted in highly contiguous and accurate plant reference genomes.
In conjunction with the work shown here, we also applied the latest chemistry and basecalling improvements to melon and lettuce. This work was presented earlier at NCM 2021. For details on these datasets please see the blog post of KeyGene NGS expert Alexander Wittenberg on the ONT website.
Datasets and background
- Arabidopsis thaliana ecotype Columbia is a diploid plant with 2n = 10 chromosomes. It became the first plant genome to be fully sequenced based on the fact that it has a (a) small genome of ~120 Mb with a simple structure having few repeated sequences (b) short generation time of six weeks from seed germination to seed set, and (c) produces large number of seeds. The sequencing was done by an international collaboration collectively termed the Arabidopsis Genome Initiative and published in 2000 (1). The Arabidopsis Information Resource (TAIR) maintains a database of genetic and molecular data for this model plant species. The latest genome assembly release of the consortium is TAIR10.1 (2). Furthermore, in August 2021 a high-quality and near complete Col-0 genome assembly (Col-XJTU) was published using a combination of Oxford Nanopore Technology ultra-long reads, PacBio HiFi reads, and Hi-C data (3). Finally, in November 2021 Naish et al. published a HiFi and ONT-based assembly (Col-CEN) that resolved all five centromeres (4). Here, we generated a dataset using only one MinION cell with the latest R10.4 pore type and Q20+ chemistry.
- Tomato (Solanum lycopersicum) is the most intensively investigated Solanaceous species both in genetic and genomics studies. It is a diploid species with a haploid set of 12 chromosomes and a relative small genome (950 Mb). Tomato line Heinz 1706 (a processing tomato) is supported by the most advanced public assembly. The International Tomato Genome Sequencing Project published the first version in 2012 (5) and the latest version, SL4.0 was released in September 2019 (6). In August 2021, scientist from China published a new Heinz 1706 reference genome generated using PacBio and ONT long reads (7). Here, we generated a dataset using just two PromethION cells with the latest R10.4 pore type and Q20+ chemistry.
High molecular weight DNA was extracted from leaf material of both species and ligation-based libraries were prepared using the early access Q20+ kit. The data was basecalled using software version 21.06.13 for the MinION cell and version 21.10.8 for the two PromethION cells. Both datasets were basecalled using the super accuracy model (that has the plant-trained model incorporated). An overview of the datasets is provided in table 1. For all runs we achieved nice read length distributions (Figure 1).
Table 1: An overview of datasets released on ENA *Calculated median percentage identity using Nanoplot against latest public reference genome for respective species.
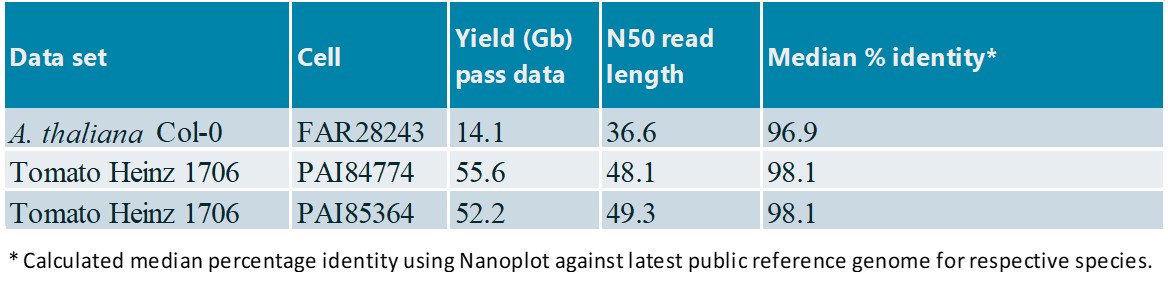
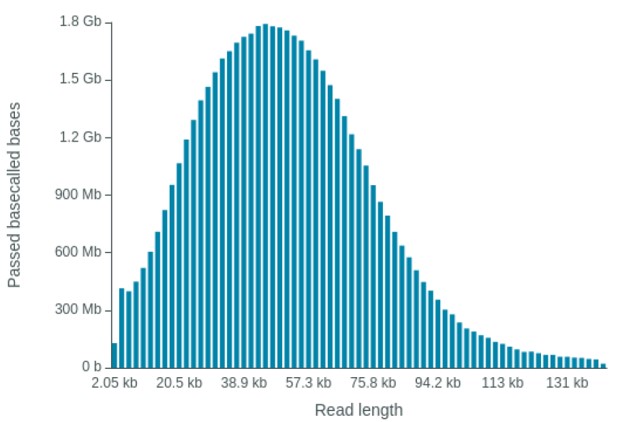
Figure 1: Read-length distribution of tomato PromethION cell PAI84774.
De novo crop genome assembly approach
De novo assemblies were generated using the publicly available Flye V2.9 assembler (8) and the KeyGene proprietary STL assembler. Consensus calling on the assemblies was performed using Medaka version 1.5.0.
Determination of consensus accuracy has been performed using Merqury version 1.3 (9) using publicly available Illumina short-read datasets.
- Arabidopsis thaliana Col-0: JGI Project ID: 1119135 (21 Gb with length 101 bp)
- Tomato Heinz SRA SRX118405 (40 Gb with paired-end length 101 bp)
The Benchmarking Universal Single-Copy Orthologs (BUSCO) gene set (10) was used to gauge the accuracy and completeness of the tomato assemblies.
A. thaliana Col-0 contig assembly results
For A. thaliana Col-0 we selected the longest 40X reads and performed Flye and STL de novo assemblies (Table 2).
Table 2: Statistic for public and KeyGene de novo assemblies A. thaliana Col-0
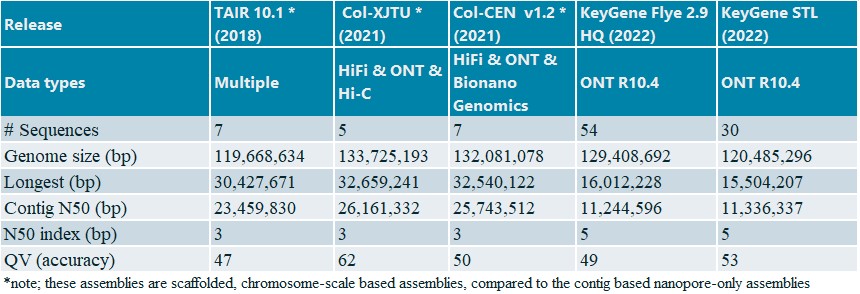
The KeyGene contig-based assemblies generated with Flye and STL show a high level of contiguity, consensus accuracy and collinearity (Figure 2) compared to TAIR v10.1 and the other two reference crop genome assemblies. Important to note: these results were obtained using just a single MinION R10.4 flow cell (no re-loading) in combination with a plant-trained basecaller and without any scaffolding.
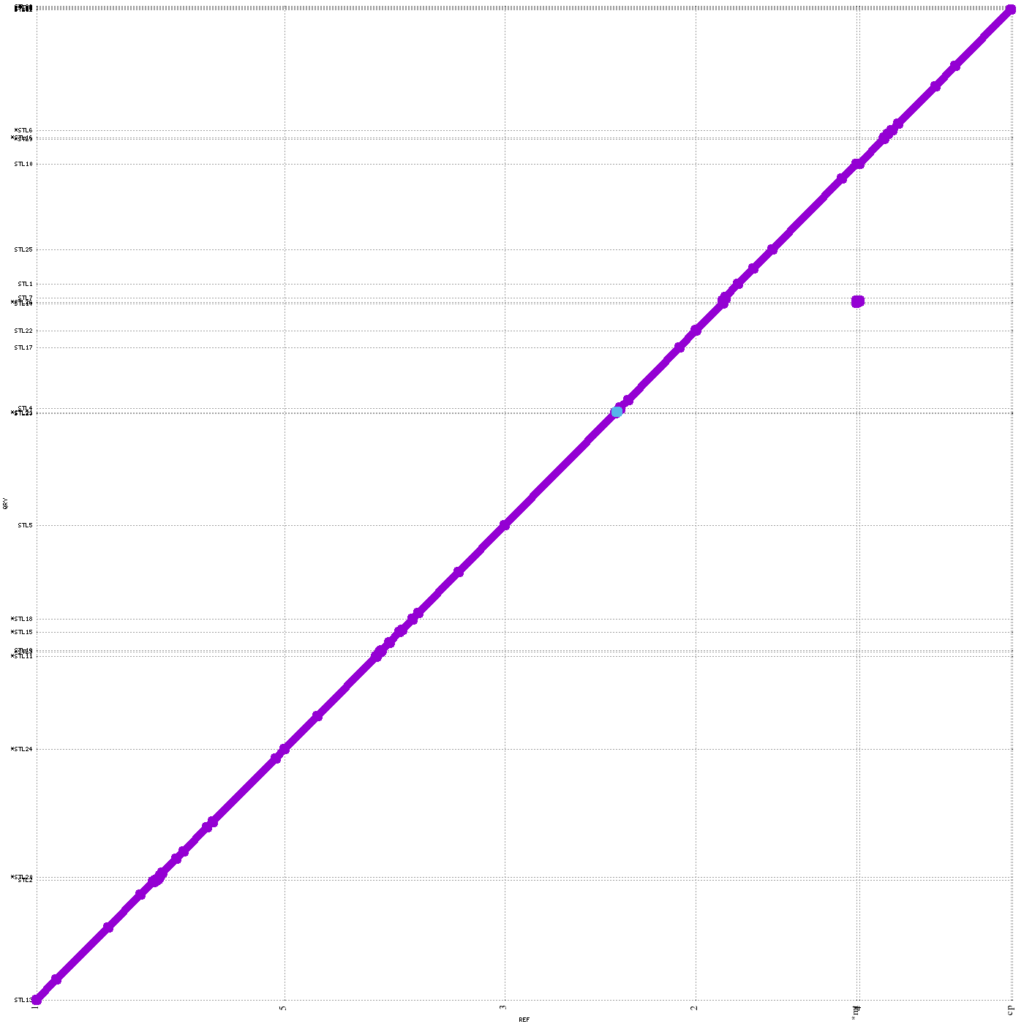
Figure 2: MUMmerplot (Marçais et al. 2018) comparison of TAIR10.1 reference genome on the X-axis with the KeyGene de novo assembled STL contig assembly on the Y-axis. The contigs were aligned using STL and the alignments were filtered for unique matches with a 10Kb length cut-off.
ENA data links (study: PRJEB49840)
- Data (.fast5 format) accession number: ERS9766930
- Flye-based assembly, accession number GCA_927333615
For tomato Heinz 1706, we performed Flye and STL de novo assemblies by selection of the longest 40X and 30X reads, respectively (Table 3).
Tomato Heinz 1706 contig assembly results
For tomato Heinz 1706, we performed Flye and STL de novo assemblies by selection of the longest 40X and 30X reads, respectively (Table 3).
Table 3: Contig statistics for public and KeyGene de novo assemblies tomato Heinz 1706
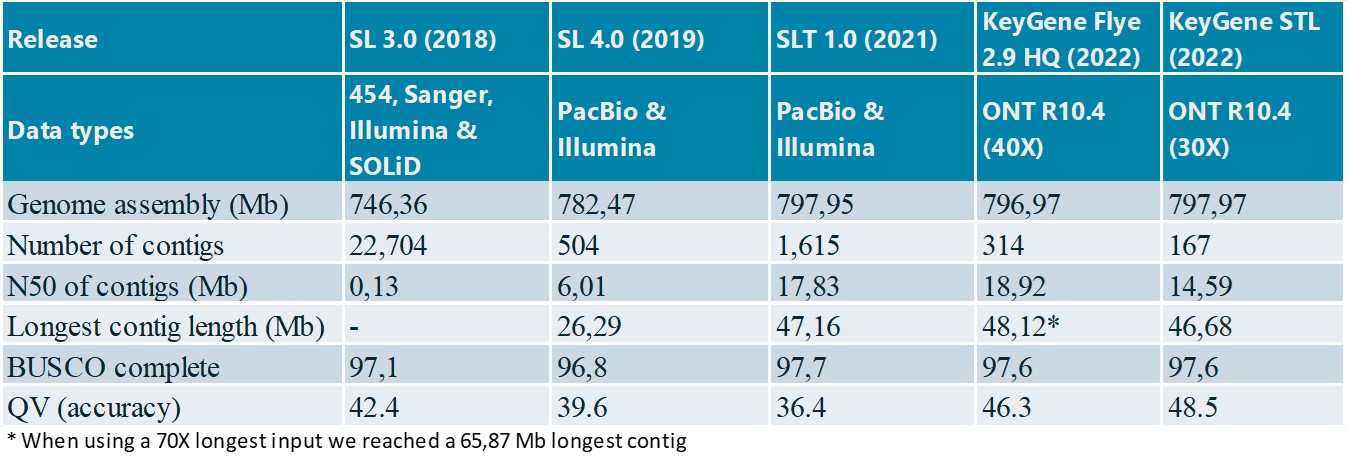
The KeyGene contig-based assemblies show better contiguity and a good collinearity with the SL4.0 public reference (Figure 3). Total assembly size and consensus accuracies for Nanopore-only assemblies are significantly higher compared to public assemblies SL 3.0 and 4.0. Noteworthy, these results were obtained using only two R10.4 PromethION flow cells in combination with a plant-trained basecaller and without short-read polishing and scaffolding. This is in contrast to the public assemblies that required a much higher coverage and at least two sequencing platforms as input for the assemblies.
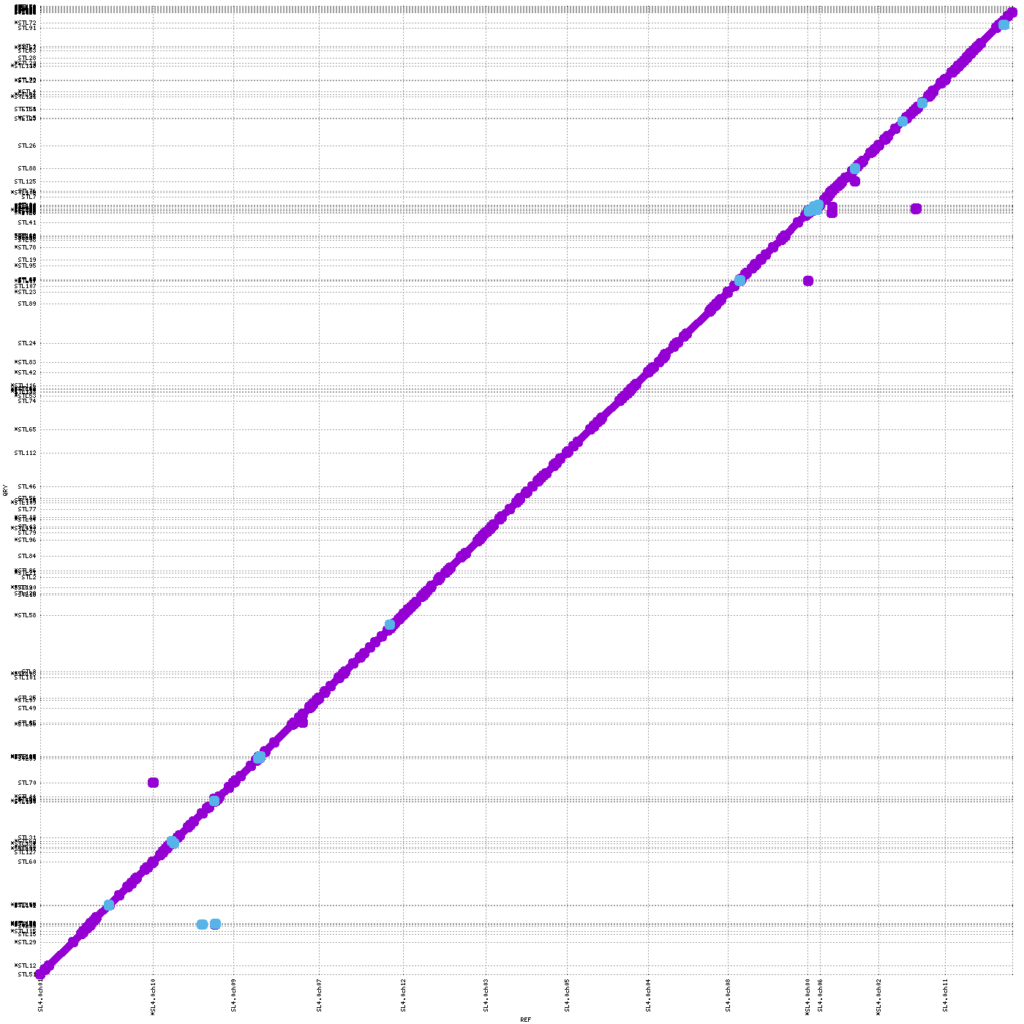
Figure 3: MUMmerplot (Marçais et al. 2018) comparison of public SL4.0 tomato reference genome on the X-axis with the KeyGene de novo assembled STL contig assembly on the Y-axis. The contigs were aligned using STL and the alignments were filtered for unique matches with a 10Kb length cut-off.
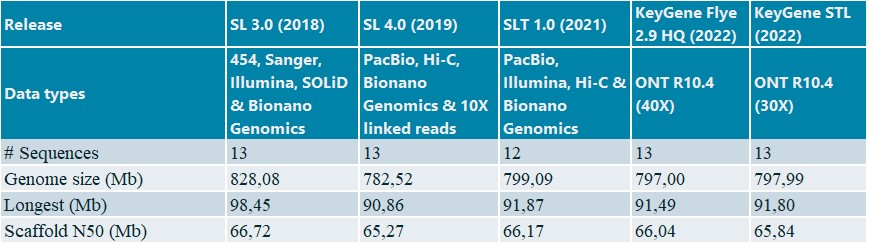
We also generated a chromosome-scale reference using RagTag (11) with assembly SL 4.0 as a guide for the KeyGene in-house assemblies and compared them against the public assemblies (Table 4).
Table 4: Scaffold statistics for public and KeyGene de novo assemblies tomato Heinz 1706
ENA data links (study: PRJEB49840)
- Data (.fast5 format) accession number: ERS9766931
- Flye-based assembly, accession number GCA_927333815.1
 Conclusion & perspectives
Conclusion & perspectives
We generated and released datasets and assemblies of two plant species based on the latest Oxford Nanopore technology and chemistry, that are comparable or better than public releases but with less effort in data generation and computational time. Data generation and analysis were completed within a week after high molecular weight DNA isolation. The KeyGene STL de novo assembler is significantly faster compared to most state-of-the-art assemblers. This is in strong contrast to public, consortium-based assemblies that have taken many years and large investments to be completed.
Raw read accuracies of Oxford nanopore reads have significantly increased in the past few months using the latest chemistry and R10.4 pore type in combination with a plant-trained basecalling model. The accuracies obtained are now in the range of 97-99% for plant species. We expect to further fine-tune the plant-trained basecalling model in the near future using data from a range of crop species. Consensus accuracies using nanopore-only data are now reaching QV ~47-53 (i.e. ~1 error/100Kb), which is exceptionally high.
These highly contiguous and accurate references have been generated with Nanopore-only data. Consequently, this will strongly reduce costs in such a way that more and more crop species can be supported by high quality reference genome sequences. Here we have shown that for a genome like tomato, a high-quality reference can already be generated for a few thousand Euros.
References
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815 (2000). https://doi.org/10.1038/35048692
- TAIR 10.1 reference assembly and The Arabidopsis Information Resource; https://www.arabidopsis.org/index.jsp
- High-quality Arabidopsis thaliana Genome Assembly with Nanopore and HiFi Long Reads. Bo Wang, Xiaofei Yang, Yanyan Jia, Yu Xu, Peng Jia, Ningxin Dang, Songbo Wang, Tun Xu, Xixi Zhao, Shenghan Gao, Quanbin Dong, Kai Ye, Genomics, Proteomics & Bioinformatics, 2021, https://doi.org/10.1016/j.gpb.2021.08.003
- The genetic and epigenetic landscape of the Arabidopsis centromeres. M. Naish et al., Science 374, eabi7489 (2021). https://doi.org/10.1126/science.abi7489
- The Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641 (2012). https://doi.org/10.1038/nature11119
- An improved de novo assembly and annotation of the tomato reference genome using single-molecule sequencing, Hi-C proximity ligation and optical maps. Prashant S. Hosmani, Mirella Flores-Gonzalez, Henri van de Geest, Florian Maumus, Linda V. Bakker, Elio Schijlen, Jan van Haarst, Jan Cordewener, Gabino Sanchez-Perez, Sander Peters, Zhangjun Fei, James J. Giovannoni, Lukas A. Mueller, Surya Saha bioRxiv 767764; doi: https://doi.org/10.1101/767764
- A high-continuity and annotated tomato reference genome. Xiao Su, Baoan Wang, Xiaolin Geng, Yuefan Du, Qinqin Yang, Bin Liang, Ge Meng, Qiang Gao, Sanwen Huang, Wencai Yang, Yingfang Zhu, Tao Lin bioRxiv 2021.05.04.441887; doi: https://doi.org/10.1101/2021.05.04.441887
- Mikhail Kolmogorov, Jeffrey Yuan, Yu Lin and Pavel Pevzner, “Assembly of Long Error-Prone Reads Using Repeat Graphs”, Nature Biotechnology, 2019 doi:10.1038/s41587-019-0072-8
- Rhie, A., Walenz, B.P., Koren, S. et al. Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol 21, 245 (2020). https://doi.org/10.1186/s13059-020-02134-9
- BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Simao, F.A., Waterhouse, R.M., Ioannidis, P., Kriventseva, E.V. and Zdobnov, E.M. (2015). Bioinformatics 31, 3210-3212.
- Automated assembly scaffolding elevates a new tomato system for high-throughput genome editing. Alonge, Michael, et al. bioRxiv (2021). https://doi.org/10.1101/2021.11.18.469135