LONG-READ SEQUENCING FOR STRUCTURAL VARIATION (SV) DETECTION
There is increasing evidence that structural variation (SV) in genomes can influence traits like resistance to pests and diseases, adaptation to climate change, yield and quality [1-3]. By exploring SV in your germplasm, you access the full spectrum of variation and you can better link those to your breeding lines and traits of interest.
At KeyGene, we offer an attractive package for SV detection, thus enabling breeders and scientists to gain Genome Insights in their crop of interest. For details, check below or get in touch with us keygenepr@keygene.com.
Exploiting SV
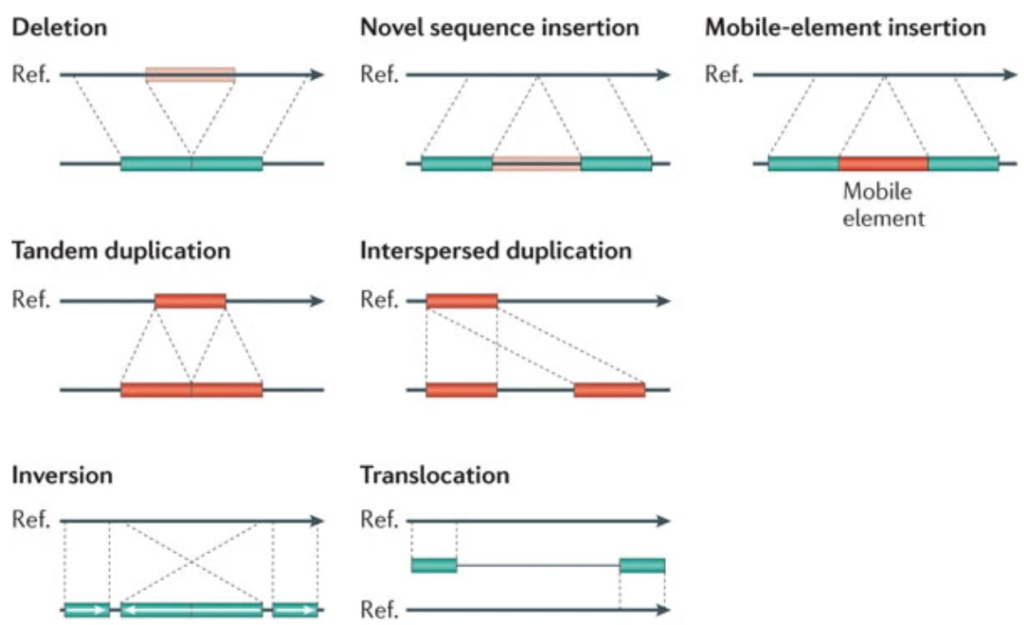
SV are genomic variations that include deletions, insertions, inversions and duplications. Having the full spectrum of variations can help breeders and plant scientists to better understand their germplasms and traits of interest.
Source image: Genome structural variation discovery and genotyping, Nature Reviews Genetics volume 12, pages 363–376(2011); Can Alkan, Bradley P. Coe & Evan E. Eichler, Nature, https://doi.org/10.1038/nrg2958
For example, instead of solely relying on Single Nucleotide Polymorphisms (SNP)-based analyses, SV can help elucidate Quantitative Trait Loci (QTL) in bi-parental populations and mine for more variations. In addition, Genome-Wide Association Studies (GWAS) using SV can help identify loci associated with traits of interest in diversity panels and germplasm collections. Furthermore, functional SV can unravel important functional gene variations.
Recently, Alonge et al. [4] detected SV underlying novel quantitative trait loci and associated with flavor and fruit weight in a diversity of 100 tomato accessions. Similarly, Liu et al. [5], identified SV associated with seed luster, flowering and iron deficiency in soybean.
Long-read sequencing for superior SV detection
Technology has been the main bottleneck to easily detect SV. Now, with advances in long-read sequencing, small to mid-size and long, complex SV can be detected [6]. At KeyGene, we use long-read sequencing on native DNA. Since no PCR step is involved, long-read sequencing results in more accurate and unbiased detection of SV, compared with short-read sequencing and other approaches. Even genome regions containing many repeats or having a high GC content are now accessible.
Our SV detection offer
 KeyGene has extensive knowledge and experience in generating Genome Insights in a wide range of crops, such as vegetables, cereals and ornamentals. We were early adopters of long-read sequencing technologies and have developed proprietary state-of-the-art analyses tools. We also have unique expertise in isolating high-quality High-Molecular-Weight plant (HMW) DNA from many crops. This combination makes us an ideal partner in obtaining and exploiting SV as a tool for Genome Insights.
KeyGene has extensive knowledge and experience in generating Genome Insights in a wide range of crops, such as vegetables, cereals and ornamentals. We were early adopters of long-read sequencing technologies and have developed proprietary state-of-the-art analyses tools. We also have unique expertise in isolating high-quality High-Molecular-Weight plant (HMW) DNA from many crops. This combination makes us an ideal partner in obtaining and exploiting SV as a tool for Genome Insights.
We offer an attractive package that includes the generation of data and analysis of SV in your crop of interest. Our offering includes:
- Oxford Nanopore Technology (ONT) 1D library preparation and barcoding
- PromethION sequencing to ~15X genome coverage
- SV analysis using an optimized pipeline with state-of-the-art tools
- Data delivery and reporting within 3-4 months from material delivery
 Of course, we offer the possibility to customize our offering to your crop and needs: We can include HMW DNA isolation, transfer raw data, deliver results in a visualization tool, increase coverage, etc.
Of course, we offer the possibility to customize our offering to your crop and needs: We can include HMW DNA isolation, transfer raw data, deliver results in a visualization tool, increase coverage, etc.
For more details, e.g. for an attractive offer for structural variation (SV) detection in your germplasm, get in contact with us: keygenepr@keygene.com
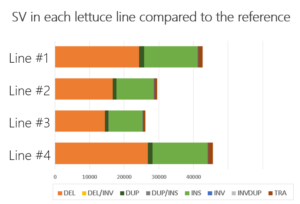
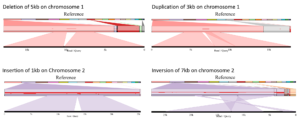
Case study: lettuce
For one of our partners, we offered insight into SV by comparing each of the four lettuce accessions with a public reference. We generated ~ 15X coverage on the PromethION for each line and used a combination of SV detection tools to analyze the data.
Our team of next-generation sequencing experts and bio-informaticians delivered an SV catalog containing more than 25,000 SV. This enabled the breeders to relate differences in genomic structures to differences in specific characteristics observed in the accessions.
Check out our short video on the perspectives of Structural variant-based genome insights.
References
Do you want to learn more about the perspectives of Genome Insights into SV? Here is a selection of articles worth reading:
- Willson, J., 2020. Resolving the roles of structural variants. Nature Reviews Genetics, pp.1-1., https://www.nature.com/articles/s41576-020-0264-6 (open access)
- Chawla, H.S., Lee, H., Gabur, I., Tamilselvan-Nattar-Amutha, S., Obermeier, C., Schiessl, S.V., Song, J.M., Liu, K., Guo, L., Parkin, I.A. and Snowdon, R.J., 2020. Long-read sequencing reveals widespread intragenic structural variants in a recent allopolyploid crop plant. bioRxiv., https://www.biorxiv.org/content/10.1101/2020.01.27.915470v1.full.pdf(open access)
- Schiessl, S.V., Katche, E., Ihien, E., Chawla, H.S. and Mason, A.S., 2019. The role of genomic structural variation in the genetic improvement of polyploid crops. The Crop Journal, 7(2), pp.127-140., https://www.sciencedirect.com/science/article/pii/S2214514118301028 (open access)
- Alonge, M., Wang, X., Benoit, M., Soyk, S., Pereira, L., Zhang, L., Suresh, H., Ramakrishnan, S., Maumus, F., Ciren, D. and Levy, Y., 2020. Major Impacts of widespread structural variation on gene expression and crop improvement in tomato. Cell, 182(1), pp.145-161., https://www.sciencedirect.com/science/article/abs/pii/S0092867420306164
- Liu, Y., Du, H., Li, P., Shen, Y., Peng, H., Liu, S., Zhou, G.A., Zhang, H., Liu, Z., Shi, M. and Huang, X., 2020. Pan-genome of wild and cultivated soybeans. Cell, 182(1), pp.162-176., https://www.sciencedirect.com/science/article/abs/pii/S0092867420306188
- Ho, S.S., Urban, A.E. and Mills, R.E., 2019. Structural variation in the sequencing era. Nature Reviews Genetics, pp.1-19., https://www.nature.com/articles/s41576-019-0180-9