4 April 2022
Maize B73 Oxford Nanopore duplex sequence data release
With the recent advances in Oxford Nanopore Technologies pore type, chemistry, and a plant-trained basecaller, we would like to announce the release of a new maize B73 dataset. The release encompasses the pass-filter duplex data (.fast5 format) together with associated reference genome assemblies.
=> KeyGene’s René Hogers will present the poster “Impact of nanopore sequencing innovations on comprehensive genomic and genetic understanding of crops” at the AGBT Ag meeting in San Diego on 6 April 2022, 10:40-12:10 local time (poster #805).
=> Read the 10 January NGS post: “Fast, contiguous and accurate arabidopsis (Col-0) and tomato (Heinz 1706) genome assembly, thanks to new chemistry, nanopores and plant-aware basecaller”
Background
 Maize is one of the most important food crops worldwide, yet accompanied by a complex, large, repetitive genome. The public consortium published the first reference of cultivar B73 in 2009 (1), while the most recent public reference Zm-B73-REFERENCE-NAM-5.0 was based on PacBio CLR reads and released in 2020 (2). Soon after, a PacBio HiFi dataset with associated assembly was released publicly (3 and 4).
Maize is one of the most important food crops worldwide, yet accompanied by a complex, large, repetitive genome. The public consortium published the first reference of cultivar B73 in 2009 (1), while the most recent public reference Zm-B73-REFERENCE-NAM-5.0 was based on PacBio CLR reads and released in 2020 (2). Soon after, a PacBio HiFi dataset with associated assembly was released publicly (3 and 4).
Since plant DNA differs in composition and base modifications significantly from human and microbial species, Oxford Nanopore Technologies embarked on training their latest basecaller with maize B73 sequencing data to achieve highest raw read accuracies for such complex plant genomes. Here we evaluated these basecalling improvements together with the impact of maize duplex reads, which consist of combined forward- and reverse strand sequence information from a single DNA molecule (see figure 1) on genome assembly of B73. Moreover, sequence data was generated by applying the latest advances of the early access R10.4 pores in conjunction with Q20+ chemistry.
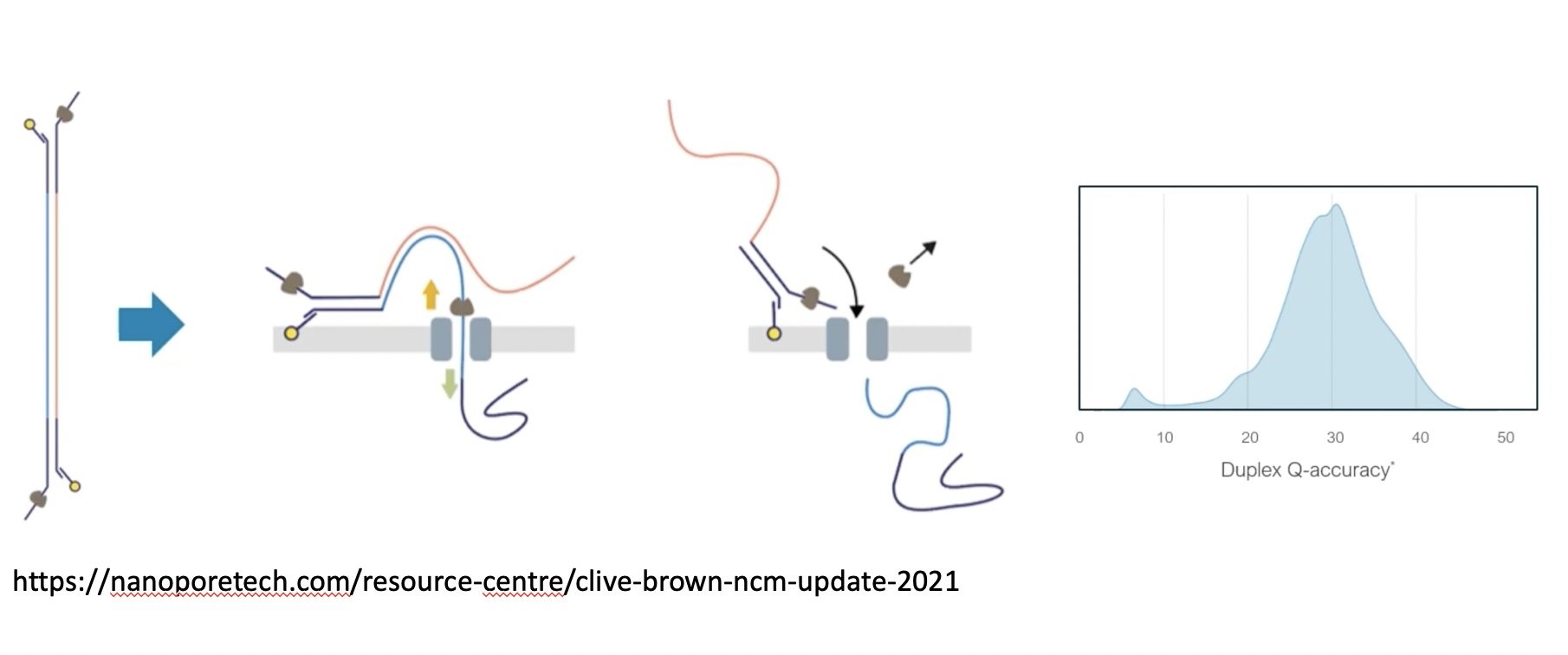
Figure 1: the principle of nanopore duplex reads resulting in raw read accuracies approaching Q30 accuracy. Source of picture
Methods
We generated a large dataset for maize B73 in collaboration with Oxford Nanopore Technologies as follows. High molecular weight nuclear DNA was extracted from young leaf material (15-20 days after sowing) and ligation-based libraries were prepared using the early access SQK-LSK112 Q20+ kit. We run twenty two flow cells on a PromethION P24 (software version 21.10.8) in combination with super accuracy basecalling. Important to note: the plant-trained basecalling model was used during super accuracy basecalling. For the duplex basecalling duplex tools v0.2.7 was applied in conjunction with Guppy v6.0.0 . Duplex reads were filtered on length (>10 Kb) prior to assembly. An overview of the datasets is provided in table 1.
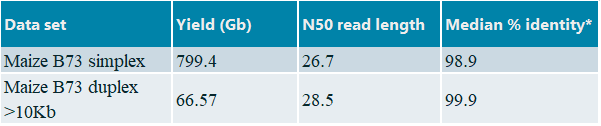
Table1: Overview of datasets, *Calculated median percentage identity using Nanoplot against NAM v5.0 public reference genome
We assembled the duplex data using the latest versions of Hifiasm (4) and HiCanu (5) using default settings. In addition to the latest NAM versions 5.0 assembly generated from 83X PacBio CLR data we also generated a new HiFi-based assembly using the public data and the latest version of Hifiasm. Using the same public HiFi data a better assembly was obtained compared to as described by Cheng et al. (4). Finally, we determined the consensus accuracy of all assemblies using Merqury (6) in combination with public Illumina dataset ENA accession no. SRR2960981
Results
Using the duplex reads only we achieved highly contiguous and accurate de novo assemblies using Hifiasm and HiCanu (Table 2).
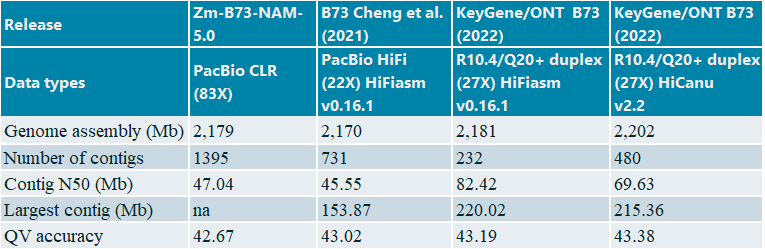
Table2: Assembly statistics of maize B73 line
The KeyGene contig-based assemblies show significantly increased contiguity: contig number goes down from 1395 and 731 to 480 and 232 depending on dataset type and assembly strategy. Furthermore, high collinearity was obtained with the NAM-5.0 public reference, as clearly shown by the almost perfectly formed diagonal in Figure 2. Also, total assembly size and consensus accuracies for Nanopore-duplex only assemblies are somewhat higher when compared to the PacBio CLR and HiFi based assembles.
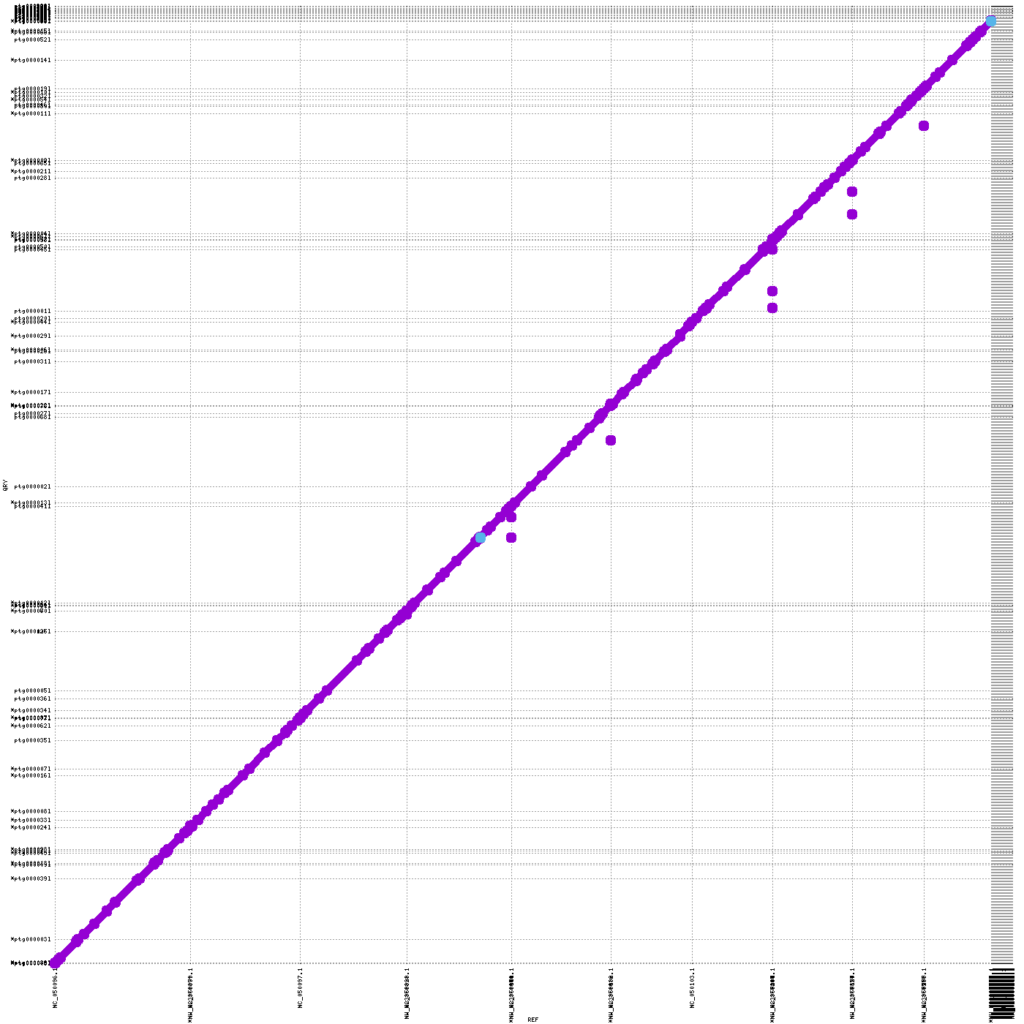
Figure 2: MUMmerplot (Marçais et al. 2018) comparison of B73 NAM version 5.0 public reference genome on the X-axis with the KeyGene nanopore duplex only hifiasm contig assembly on the Y-axis. The contigs were aligned using an aligner internally developed byKeyGene; the alignments were filtered for unique matches with a size cut-off of 10Kb.
ENA data links (study: PRJEB49840)
Data (.fast5 format) accession number: ERR9463595
Hifiasm-based assembly, accession number: GCA_933823745.2 > will be made available soon
HiCanu-based assembly, accession number: GCA_933823745.3 > will be made available soon
As stated above, the Oxford Nanopore Technologies PromethION duplex data applied here was generated using ~10x more flow cells when compared to PacBio Sequel IIe generated HiFi data. However, increased data output and accuracy with a novel, fast Q20+ chemistry will fill the yield gap and is planned to be evaluated in early access during Q2 2022.
More information on the Q20+ chemistry and how it is being used by the community can be found here.
References
- The B73 maize genome: complexity, diversity, and dynamics. 2009 Nov 20;326(5956):1112-5. doi: 10.1126/science.1178534
- https://www.ncbi.nlm.nih.gov/assembly/GCF_902167145.1/ and NAM genomes; https://nam-genomes.org/
- Hon, T., Mars, K., Young, G. et al. Highly accurate long-read HiFi sequencing data for five complex genomes. Sci Data 7, 399 (2020). https://doi.org/10.1038/s41597-020-00743-4
- Cheng, H., Concepcion, G.T., Feng, X., Zhang, H., Li H. (2021) Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat Methods, 18:170-175. https://doi.org/10.1038/s41592-020-01056-5 and https://github.com/chhylp123/hifiasm
- Nurk S, Walenz BP, Rhiea A, Vollger MR, Logsdon GA, Grothe R, Miga KH, Eichler EE, Phillippy AM, Koren S. HiCanu: accurate assembly of segmental duplications, satellites, and allelic variants from high-fidelity long reads. biorXiv. (2020). doi: 1101/gr.263566.120
- Rhie, A., Walenz, B.P., Koren, S. et al. Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol 21, 245 (2020). https://doi.org/10.1186/s13059-020-02134-9
Contact
 Would you like to learn more about recent developments at KeyGene in technology innovation in the field of Next Generation Sequencing? Send a mail to Alexander Wittenberg? Send a mail to Alexander Wittenberg
Would you like to learn more about recent developments at KeyGene in technology innovation in the field of Next Generation Sequencing? Send a mail to Alexander Wittenberg? Send a mail to Alexander Wittenberg