19 November 2021
Genome of ‘Mother of Sour Cherries’ decoded and published
 An international team of scientists from KeyGene, the Julius Kühn-Institute for Fruit Breeding Research in Germany; Szent István University in Hungary, the Hungarian University of Agriculture and Life Science, the University of Greifswald, and the University of Hohenheim, both in Germany, have recently succeeded in decoding the building blocks of the genome of the ground cherry, Prunus fruticosa. The team published their results in Genomics .
An international team of scientists from KeyGene, the Julius Kühn-Institute for Fruit Breeding Research in Germany; Szent István University in Hungary, the Hungarian University of Agriculture and Life Science, the University of Greifswald, and the University of Hohenheim, both in Germany, have recently succeeded in decoding the building blocks of the genome of the ground cherry, Prunus fruticosa. The team published their results in Genomics .
Latest technology
Thanks to the strategic collaboration between KeyGene and Oxford Nanopore Technologies, the team had access to technology not yet on the market for DNA-sample preparation (the LSK110 library preparation kit) and DNA sequencing (the R10.3 flow cell). The KeyGene team was also responsible for the bioinformatics analysis of the genome.
The research team was thus able to decode the largest (tetraploid) cherry genome to date, with a length of 1.1 billion bases, covering 4 * 8 chromosomes. The 8 chromosomes derived from the analysis, the ‘haploid’ set, add up to a total length of 366 million bases.
Innovation for breeding
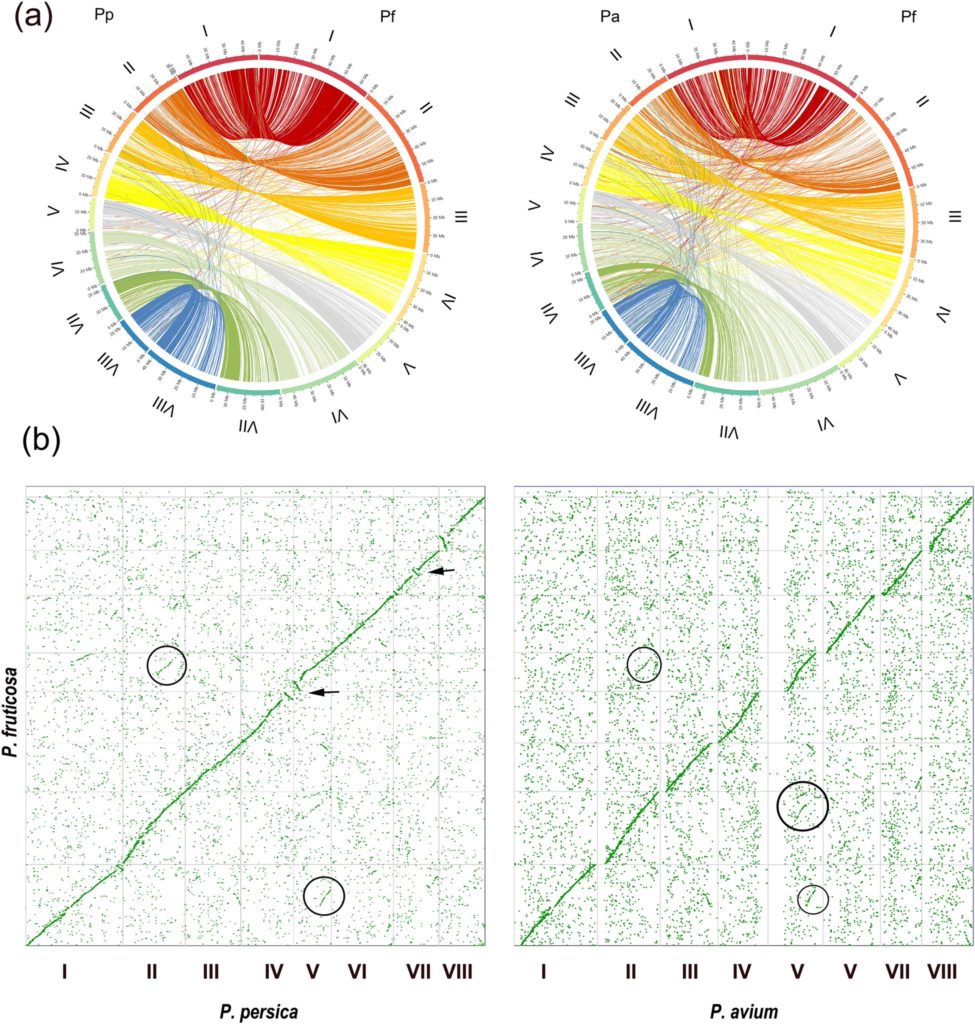 With the first genome of the ground cherry now decoded, it is possible to better breed improved cherry varieties and understand the evolutionary development of the sour cherry. Based on the genetic code, the researchers can now assign genes of the ground cherry in the genome of the sour cherry. This makes it possible to make better predictions about positive or negative traits when breeding new sour cherry varieties. With the help of this knowledge, breeders can make the sour cherry more robust against diseases and fit for climate change.
With the first genome of the ground cherry now decoded, it is possible to better breed improved cherry varieties and understand the evolutionary development of the sour cherry. Based on the genetic code, the researchers can now assign genes of the ground cherry in the genome of the sour cherry. This makes it possible to make better predictions about positive or negative traits when breeding new sour cherry varieties. With the help of this knowledge, breeders can make the sour cherry more robust against diseases and fit for climate change.
Sour cherry: descendant from the ground cherry and the sweet cherry
The sour cherry (Prunus cerasus L.) is traditionally found in almost every allotment garden. Whether on a cake, as jam or juice, the fruit enjoys great popularity. Sour cherries originated in Eastern Europe and Asia Minor (Anatolia). Their parents are the sweet cherry (P. avium), which is widespread in Europe and Asia Minor, and the ground cherry (P. fruticosa), which is common in the vast steppes of Eastern Europe and Western Asia.
As early as the late 1960s, scientists succeeded in proving for the first time that the sour cherry is a natural hybrid between the ground cherry and the sweet cherry. While the two parental species initially evolved independently of each other, hybridization later occurred by chance in areas where both species occur at the same time, which then gave rise to our present-day sour cherries.
The genome of the sour cherry, therefore, consists of two parts. One-half of the chromosomes originates from the sweet cherry, while the other half originates from the ground cherry. The genome of the sweet cherry has already been sequenced. So far, such a genome sequence is missing for the ground cherry. In order to draw conclusions about the structure of the sour cherry genome, the now developed knowledge about the genome sequence of the ground cherry is very helpful.
 Read the full (open access) paper in Genomics:
Read the full (open access) paper in Genomics:
Wöhner, T., Emeriewen O., Wittenberg A., et. al.: The draft chromosome-level genome assembly of tetraploid ground cherry (Prunus fruticosa Pall.) from long reads. Genomics 113 (2021), 4173-4183.
Read more about KeyGene’s work on genome insights for crop improvement, or contact us, to discover the perspectives of partnering with KeyGene.